Solvation of Biomolecules
Water as the ubiquitous solvent of biomolecular systems plays important roles for biological function, as biomolecules have evolved and optimized their specific functions under aqueous solvation conditions. However, beyond structure formation, as e.g. in protein folding or the self-assembly of amphiphilic molecules into higher-order structures such as bilayers or micelles, the precise role of water molecules for biomolecular function often remain enigmatic at the atomic level.
Recently, we investigated a prototypical peripheral membrane protein, annexin B12, and analyzed how exactly solvent properties are affected due to the presence of i) the lipid membrane, and ii) the protein itself (see Figure). Our molecular dynamics simulations enable one to relate changes in the dynamics of these water molecules, as experimentally derived from Overhauser dynamic nuclear polarization (ODNP)-enhanced NMR relaxometry, to thermodynamic properties that are not directly accessible to experiment, such as solvent entropy. The altered properties of the solvent could have implications for molecular recognition close to membrane interfaces, e.g., the formation of protein-protein complexes.
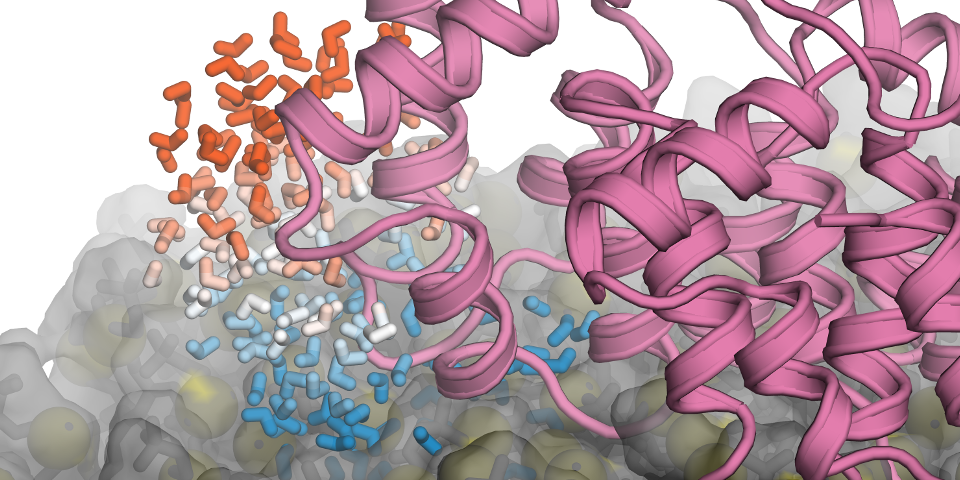
Snapshot from atomistic molecular dynamics simulation of the annexin B12 protein bound to a phospholipid membrane. A few selected water molecules in the vicinity of the protein are color-coded according to the retardation of their dynamics, from blue (slow waters) to red (water dynamics as fast as in bulk water). The observed retarded water dynamics is linked to a corresponding reduction of water entropy, with water molecules that are close to the lipid membrane being most strongly affected.