Collective Motions of Solvated Biomolecules
Liquid water forms extended hydrogen-bonded networks. In the hydration shells of a biomolecule, water motion usually slows down compared to bulk and the network stiffens. Inside a typical biologicalcell, biomolecules are tightly packed (crowded), with molecular surfaces separated by hydration layers of only around 10 Å thickness.
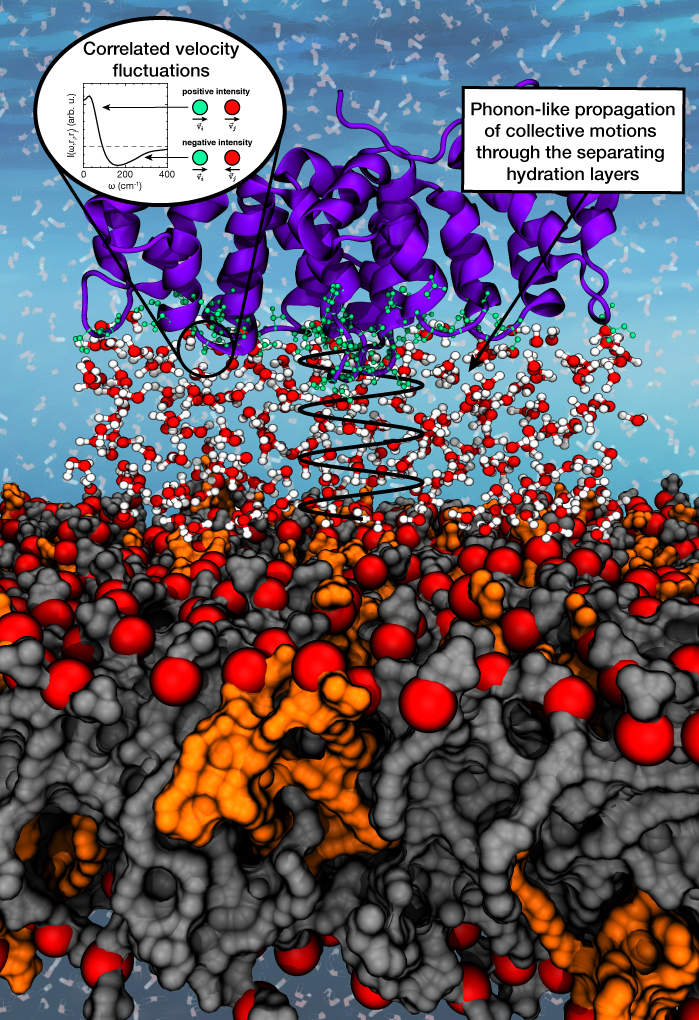
The peripheral membrane protein annexin B12 close to a phospholipid bilayer. The separating layers of hydration water mediate correlated motions between the protein and the lipid headgroups. How far do the collective motions persist?
It is therefore desirable to understand whether, and if so how, the correlated water dynamics within the hydrogen-bonded networks couple to the motions of the biomolecules, especially in crowded solutions such as the cytosol. We use molecular dynamics simulations to study the collective motions between biomolecules and their water environment based on velocity fluctuations. The identified acoustic modes may be a general mechanism for biomolecules to efficiently dissipate excess thermal energy.