|
|
|
|
CPMD Tutorial Part 7 |
|
8. Exercise: Bulk Systems
![]()
8.1. Bulk Silicon
![]()
8.2. Hydronium Ion in Bulk Water
![]()
![]() Part 6
Part 6
![]() Part 8
Part 8
![]() Start
Start
![]() Contents
Contents
 After we have (hopefully) become acquainted with running CPMD jobs and
Car-Parrinello MD runs for some systems, that are easy to follow and set
up, we can now look into bulk systems, where using a plane wave basis
set becomes a big advantage. Again we start with a rather simple system,
bulk silicon, to explore the various options and look into some
idiosyncrasies of CPMD calculations.
After we have (hopefully) become acquainted with running CPMD jobs and
Car-Parrinello MD runs for some systems, that are easy to follow and set
up, we can now look into bulk systems, where using a plane wave basis
set becomes a big advantage. Again we start with a rather simple system,
bulk silicon, to explore the various options and look into some
idiosyncrasies of CPMD calculations.
 ... some explanations/theory needed.
... some explanations/theory needed.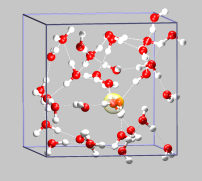
The next step is a more typical application of the CPMD code: a Car-Parrinello MD simulation of a bulk system with water. In this specific example, we try to look at the Grotthuss mechanism for proton transport in water. Our system will consist of 32 water molecules and one hydronium ion (note the CHARGE keyword in the &SYSTEM section). To speed up the equilibration phase, we start from a restart configuration (32spce-h3op-1ns.xyz) that has been equilibrated with classical MD for about 1 ns using the SPC/E water potential and an accordingly parameterized hydronium ion potential
As usual, we start with a wavefunction optimization (see 1-proton-wfopt.inp).
We want to run the MD at 400 Kelvin, so we now run a short MD with temperature rescaling for the atoms and not thermostat for the electrons (2-proton-equilib.inp).
Now we are ready to start the production run with (at least) 2000 steps. (3-proton-md.inp). Be sure to move the outputs from the equilibration out of the way before you start production.
To illustrate the structural diffusion mechanism, you can load the VMD visualization script 32h2o-h3oplus-cpmd.vmd, which will read in the TRAJEC.xyz file. The yellow line traces the position of the hydronium ion (highlighted by a transparent yellow sphere) during the trajectory. The other colored lines follow the positions of (some) of the individual protons involved in the structural diffusion process.
See elsewhere on this homepage for a more detailed version of this example.
Full Table of Contents
![]()
1. Introduction
1.1. Development Notice
1.2. Notes
1.3. Recent Changes
1.4. Citation / Bookmark
2. Table of Contents
3. Preparation and Installation Issues
3.1. Compiling CPMD
3.2. Running CPMD
3.3. Running cpmd2cube
4. The Theory: Some Fundamental Infos and Useful Literature
5. The Basics: Running CPMD, Input and Output Formats
5.1. Wavefunction Optimization: a) Input File Format
5.2. Wavefunction Optimization: b) Output File Format
5.3. Geometry Optimization
5.4. Car-Parrinello Molecular Dynamics
5.5. Further Job Types
5.6. How to Use the Tutorial
6. Exercise: Electron Structure and Geometry Optimization
6.1. Hydrogen Molecule
6.2. Water Molecule
6.3. Ammonia Molecule
7. Exercise: Car-Parrinello Molecular Dynamics
7.1. Hydrogen Molecule
7.2. Ammonia Molecule in Gas Phase
7.3. Glycine Molecule in Gas Phase
7.4. Glycine with Thermostats
8. Exercise: Bulk Systems
8.1. Bulk Silicon
8.2. Hydronium Ion in Bulk Water
9. Exercise: Determination of Dynamic Properties
9.1. Calculation of Vibrational Spectra
9.2. The 'Dragging Effect'
10. Proton Transfer in a Catalytic Triade Model
10.1. Preparing a Model from a Large System
10.2. Equilibration with a Blocked Reaction Path
10.3. Modelling Part of the Reaction Path
10.4. Calculating Electron Structure Properties and Visualizations
11. Credits
12. Downloads
13. File distribution policy
|
|
|
||
|
|||